This video shows how to apply the new spaCy v3 features as we work our way through implementing a new custom component from scratch. We build a machine learning model in Thinc, implement a new spaCy component, train it with the new configuration system and demonstrate how to use a pre-trained transformer model from the Hugging Face Transformers library to boost your performance.
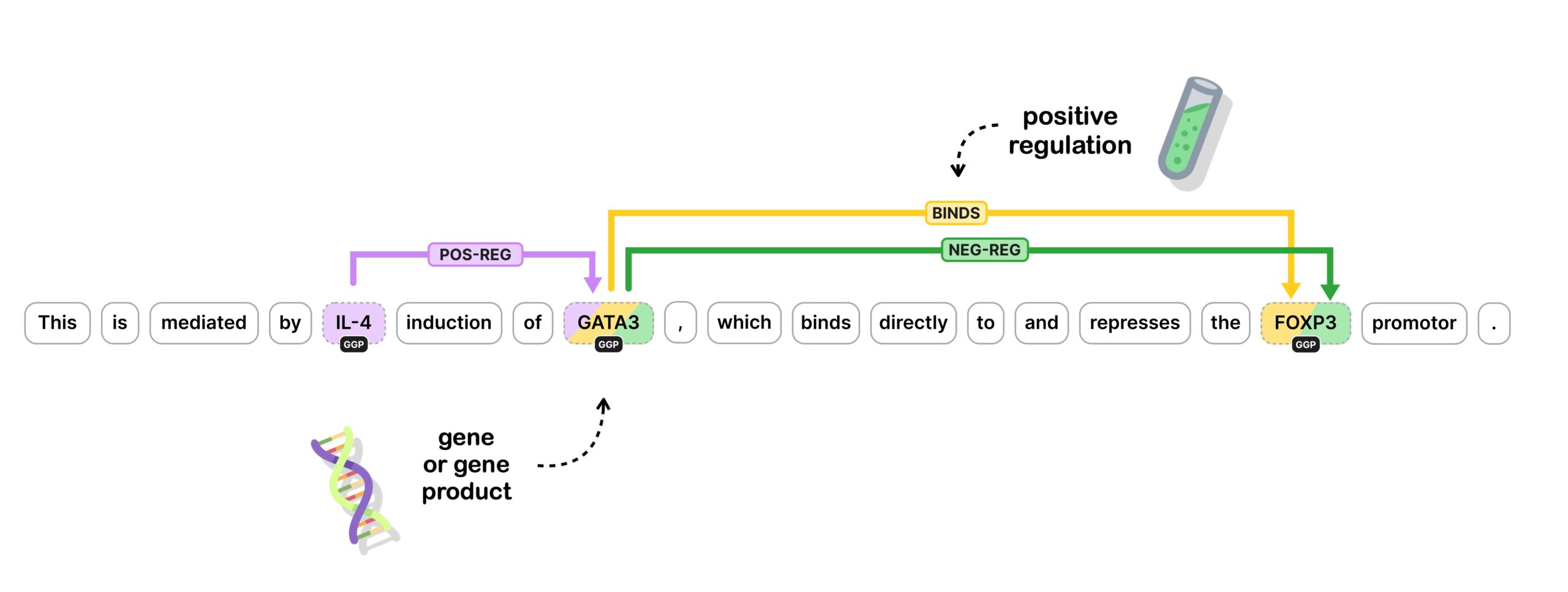
The specific challenge we are setting ourselves here is implementing a custom component to predict relationships between named entities, also called relation extraction. In the most basic form, we take two entities previously predicted by a named entity recognizer and try to determine whether there is a semantic relationship between them and, if so, label it.
In this video, I focus on predicting biomedical relations between genes and proteins. Biomedical NLP is a research area that I am passionate about, and I’ve worked in this domain extensively during my PhD and postdoc, now many years ago. For demonstration purposes, I’ve simplified the challenge and the annotation format quite a bit.
You can adapt this approach to predict any type of relations between any type of entities. The performance of your relation extraction module will depend on the specific challenge and dataset, but keep in mind that predicting relations from text is a difficult task overall.
→ Authors: Sofie Van Landeghem, Ines Montani
→ Video: Youtube
→ Code: Github
→ Transcription: Blog post