Motivation:
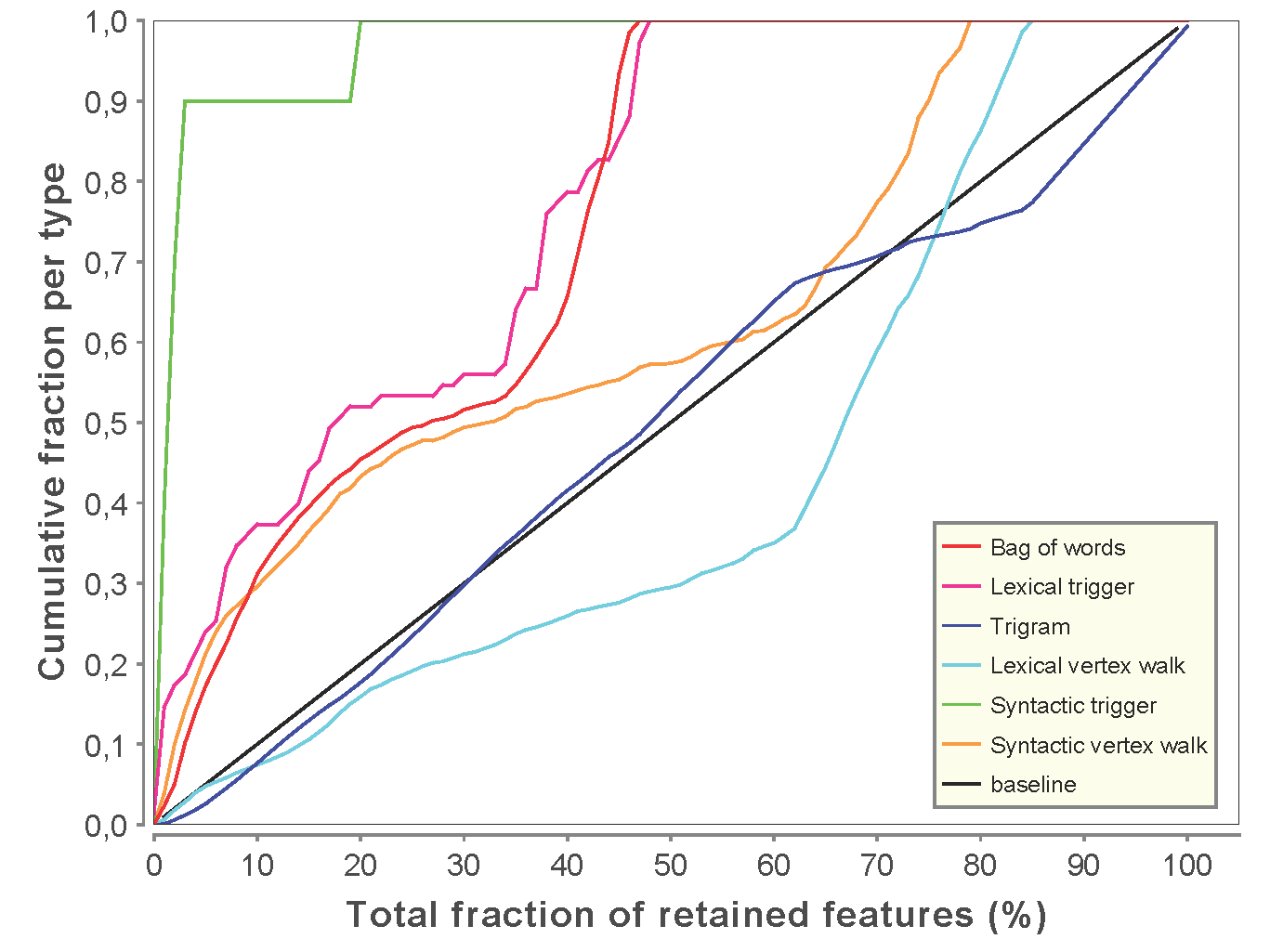
In the field of biomolecular text mining, black box behavior of machine learning systems currently limits understanding of the true nature of the predictions. However, feature selection (FS) is capable of identifying the most relevant features in any supervised learning setting, providing insight into the specific properties of the classification algorithm. This allows us to build more accurate classifiers while at the same time bridging the gap between the black box behavior and the end-user who has to interpret the results.
Results:
We show that our FS methodology successfully discards a large fraction of machine-generated features, improving classification performance of state-of-the-art text mining algorithms. Furthermore, we illustrate how FS can be applied to gain understanding in the predictions of a framework for biomolecular event extraction from text. We include numerous examples of highly discriminative features that model either biological reality or common linguistic constructs. Finally, we discuss a number of insights from our FS analyses that will provide the opportunity to considerably improve upon current text mining tools.
→ Paper: Bioinformatics
→ Authors: Sofie Van Landeghem, Thomas Abeel, Yvan Saeys, Yves Van de Peer