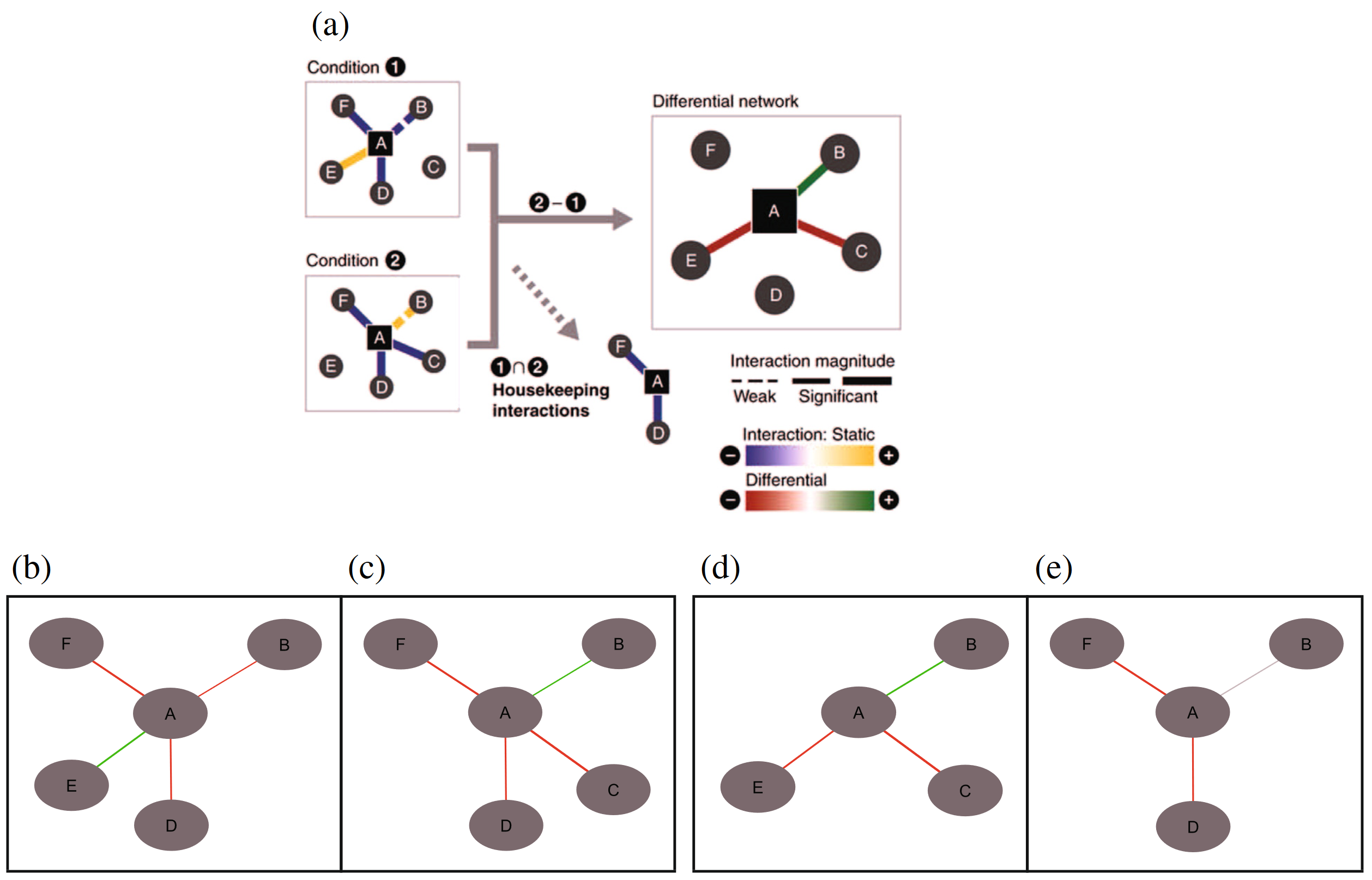
Background:
Differential networks have recently been introduced as a powerful way to study the dynamic rewiring capabilities of an interactome in response to changing environmental conditions or stimuli. Currently, such differential networks are generated and visualised using ad hoc methods, and are often limited to the analysis of only one condition-specific response or one interaction type at a time.
Results:
In this work, we present a generic, ontology-driven framework to infer, visualise and analyse an arbitrary set of condition-specific responses against one reference network. To this end, we have implemented novel ontology-based algorithms that can process highly heterogeneous networks, accounting for both physical interactions and regulatory associations, symmetric and directed edges, edge weights and negation. We propose this integrative framework as a standardised methodology that allows a unified view on differential networks and promotes comparability between differential network studies. As an illustrative application, we demonstrate its usefulness on a plant abiotic stress study and we experimentally confirmed a predicted regulator.
We made the algorithms available as an open-source package, a commandline interface and a cytoscape plugin.
→ Paper: BMC Bioinformatics
→ Code: Github repo
→ Authors: Sofie Van Landeghem, Thomas Van Parys, Marieke Dubois, Dirk Inzé, Yves Van de Peer